Chromatography Quiz #22 Results
Pickering Labs would like to congratulate our winners of our last newsletter’s AAA quiz: Narjes Ghafoori from LA County Environmental Toxicology Lab, Joy Gottlieb from New Mexico Department of Health Scientific Lab Division, Tom Schneider from Suffolk County Water Authority and Helene Lachance from Shur-Gain Nutreco!
 They have each won and will shortly be receiving: a Picnic Basket Gift Tin from Harry & David! From their webpage: Celebrate the season with this gourmet gift basket, featuring a wonderful assortment of snacks, including fruit, meat, cheese, and crackers perfect for picnics and lunches in the park. In addition to our remarkably juicy Royal Verano Pears, we're offering a range of picnic delights, like white cheddar cheese, hickory smoked summer sausage, peanut butter pretzels, sweet raspberry galettes, and more. Packaged in an exclusively designed picnic-ready tin, this gift is ready to help you make any occasion a special one
They have each won and will shortly be receiving: a Picnic Basket Gift Tin from Harry & David! From their webpage: Celebrate the season with this gourmet gift basket, featuring a wonderful assortment of snacks, including fruit, meat, cheese, and crackers perfect for picnics and lunches in the park. In addition to our remarkably juicy Royal Verano Pears, we're offering a range of picnic delights, like white cheddar cheese, hickory smoked summer sausage, peanut butter pretzels, sweet raspberry galettes, and more. Packaged in an exclusively designed picnic-ready tin, this gift is ready to help you make any occasion a special one
We hope our quiz winners enjoy their prizes and the springtime weather!
Thank you all for your submissions!
The correct answer to the Carbamates Analysis quiz:
The answer for last quarter’s quiz was: incorrectly prepared reagent. We prepared our “hydrolysis reagent” with CB910 instead of CB130. The hydrolysis reagent CB130 is at a pH of 12.5, which is much more basic than our OPA diluent CB910 at a pH of 9.1. From our Carbamates Manual: The separated carbamates are first saponified by NaOH at 100°C to release an alcohol, carbonate, and methylamine. In the second post-column reaction, methylamine reacts with OPA and Thiofluor to form the highly fluorescent derivative. So, if there is insufficient NaOH present for the first reaction, some of the carbamates do not fully hydrolyze.
Chromatography Quiz #23: Polyether Antibiotics Analysis
What caused the noise for the blue signal in the troubleshooting chromatogram below? Simply email your multiple choice answer as well as your full contact information to Rebecca at rlsmith@pickeringlabs.com by July 1st, 2016 in order to win. You will receive email confirmation that your submission has been received. The answer to the quiz and winner congratulations will be published in the next issue (to be anonymous, please notify Rebecca in submission).
Polyether Antibiotics Analysis
Pinnacle PCX post-column instrument (two-pump) is being used in a traditional HPLC setup as recommended by Pickering Laboratories. The reference chromatogram and troubleshooting chromatogram are both shown.
Narasin Standard: 2.5 µg/mL, 100uL injection
Pickering Column: 2381750, Polyether Column, C18, 4.6x250mm
Normal Operating Conditions: (for reference only, condition changes may be reflected in chromatogram)
Column Temperature: 40 °C
Flow rate: 0.7 mL/min
Isocratic: 90% Methanol, 10% of 5% Acetic Acid solution in water
Post-column conditions:
Reagent 1: Concentrated Sulfuric Acid / Methanol (4:96 v/v)
Reagent 2: 60g of Vanillin in 950mL of Methanol
Reactor 1: Ambient, 0.1mL
Reactor 2: 90 °C, 1.4mL
Reagent flow rates: 0.3 mL/min
Black (Reference) Signal:
DAD detector 520nm with bandwidth of 4nm
No reference wavelength
Sampling rate >0.10min (2.0 S response time) (2.5Hz)
Blue (Troubleshooting) Signal:
DAD detector 520nm with bandwidth of 4nm
Reference wavelength of 360nm
Sampling rate >0.05 min (1.0 S response time) (5HZ)
Can you identify the error made when running the chromatogram?
Multiple Choices:
A) Bad lamp
B) Reference Wavelength
C) Sampling rate
D) All of the above
Troubleshooting:

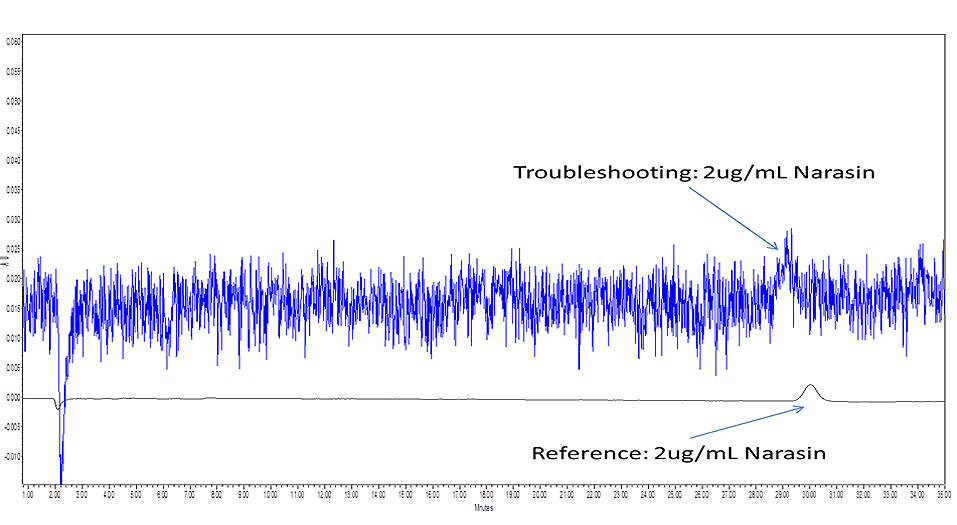

This appears to be a really bad choice of reference wavelength. The vanillin absorbs like crazy from 200-350 nm. At almost 60g/L the reference absorption will be topped out at >4AU. No DAD detector I know of can handle this. The noise is simply the remaining electronic noise from the 360 nm (plus bandwidth) diodes on the array. The solution is to move the ref wavelength out to, say, 600 nm since the antibiotics probably don't absorb there or, better, turn off the ref wavelength, acquire and store whole spectra, and do post-processing after the analysis when a clear region of the spectrum can be found.